Cutting Edge Research
Welcome to Our Computational Systems Biology Lab
We are passionate about the development and application of computational modeling methods to study the operating mechanisms of gene regulatory networks driving cellular state transitions.

Systems biology modeling of gene regulatory networks
Gene regulatory mechanisms of cell state transitions

Single cells genomics

What we do is important
We use systems biology approaches to uncover the underlying principles governing the operation of genetic networks. Specifically, we integrate computational modeling and data analysis to elucidate the relationship among robustness of network dynamics, stochasticity in gene expression and heterogeneity in a cell population. Our studies will contribute to a systems-level understanding of biological processes determining cellular state transitions.
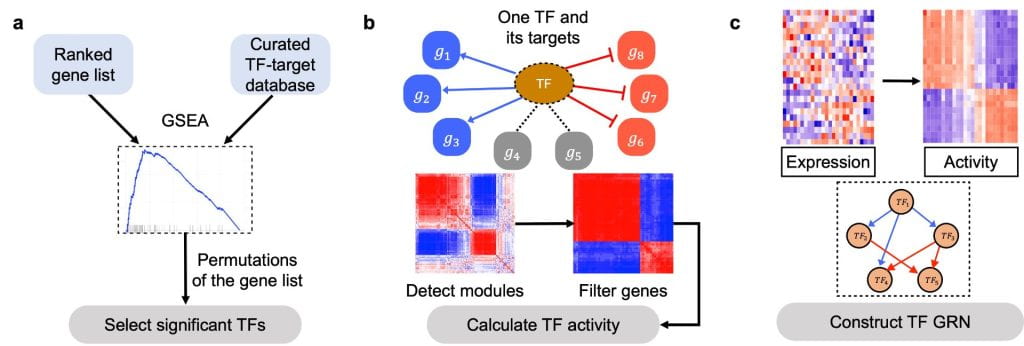
NetAct: a computational platform to construct core transcription factor regulatory networks using gene activity
A major question in systems biology is how to identify the core gene regulatory circuit that governs the decision-making process. Here, we develop a computational platform, named NetAct, for constructing core transcription factor regulatory networks using both transcriptomics data and literature-based transcription factor-target databases. NetAct robustly infers regulators’ activity using target expression, constructs network based on transcriptional activity, and integrates mathematical modeling for validation. Our in-silico benchmark test shows that NetAct outperforms existing algorithms in inferring transcriptional activity and gene networks. We illustrate the application of NetAct to model networks driving TGF-β-induced epithelial-mesenchymal transition and macrophage polarization.
Contact

m.lu@northeastern.edu